This task creates a spectral index image and calculates statistics from it and from an associated ENVIAgCrops object. It creates a classification raster showing the relative means of crops or a grayscale raster showing actual mean values of crops. For example, if the input raster is a vegetation index such as NDVI, the crop means describe the health of the crops.
ENVIAgCalculateAndRasterizeCropMetrics is a metatask that automatically runs the following tasks:
You can run AgCropCount Task or AgCountAndRasterizeCrops Task to create the ENVIAgCrops object that is required for this task. Or, you can use an ENVIAgCrops file that you created and saved earlier.
This task requires a separate license for the ENVI Crop Science Module; contact your sales representative for more information.
Example
This example performs the following steps:
- Opens an orthophoto of a citrus orchard.
- Creates a spectral index raster to make the citrus trees bright against the background. A Modified Triangular Vegetation Index (MTVI) works well in this case and will be used for crop counting. This spectral index is unrelated to the one used to calculate crop metrics.
Tip: See SpectralIndex Task for a list of spectral indices. You can run ENVIQuerySpectralIndicesTask to see what indices are available for your particular image.
- Runs ENVIAgCropCount to count the trees and to create an ENVIAgCrops object.
- Runs ENVIAgCalculateAndRasterizeCropMetricsWithSpectralIndex task, using a Normalized Difference Vegetation Index (NDVI) as the basis for calculating metrics.
- Displays a classification raster showing relative NDVI means of the citrus trees, overlaid on the orthophoto.
- Prints the range of NDVI values.
e = ENVI()
File = Filepath('CitrusOrthophoto.dat', $
Subdir=['data','crop_science'], $
Root_Dir=e.Root_Dir)
Raster = e.OpenRaster(File)
MTVIImage = ENVISpectralIndexRaster(Raster, 'MTVI')
outCrops = ENVIAgCropCount(MTVIImage, 2.0, 5.0, $
GAUSSIAN_FACTOR = 0, $
/INCLUDE_EDGES, $
INTENSITY_THRESHOLD = 0.8, $
NUMBER_OF_INCREMENTS = 4, $
PERCENT_OVERLAP = 100.0)
Task = ENVITask('AgCalculateAndRasterizeCropMetricsWithSpectralIndex')
Task.INPUT_RASTER = Raster
Task.INDEX = 'Normalized Difference Vegetation Index'
Task.INPUT_CROPS = outCrops
Task.Execute
DataColl = e.Data
DataColl.Add, Task.OUTPUT_RASTER
View = e.GetView()
Layer = View.CreateLayer(Raster)
Layer.Transparency = 25
Layer2 = View.CreateLayer(Task.OUTPUT_RASTER)
View.Zoom, /FULL_EXTENT
crops = Task.OUTPUT_CROPS
Print, 'Minimum NDVI value for entire image: ',MIN(crops.CROP_MIN)
Print, 'Maximum NDVI value for entire image: ',MAX(crops.CROP_MAX)
Syntax
Result = ENVITask('AgCalculateAndRasterizeCropMetricsWithSpectralIndex')
Input parameters (Set, Get): CLASSIFICATION, INDEX, INPUT_CROPS, INPUT_RASTER, OUTPUT_CROPS_URI, OUTPUT_METRIC, OUTPUT_RASTER_URI
Output parameters (Get only): OUTPUT_CROPS, OUTPUT_RASTER
Parameters marked as "Set" are those that you can set to specific values. You can also retrieve their current values any time. Parameters marked as "Get" are those whose values you can retrieve but not set.
Input Parameters
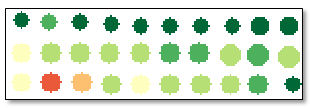
CLASSIFICATION (optional)
Set this parameter to true (the default value) to create a classification raster where crops are colored according to relative means. For example, a dark green polygon corresponds to a crop with a higher mean value than light-green, yellow, and red polygons. The resulting raster will look similar to the following:
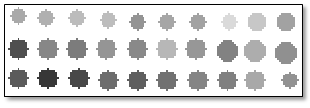
If set to false, the output will be a grayscale raster that is based on the actual mean values of the crops; for example:
INDEX (required)
Specify a string indicating the spectral index from which to calculate crop metrics. See SpectralIndex Task for a list of spectral indices. You can also run QuerySpectralIndices Task to see what indices are available for your particular image.
INPUT_CROPS (required)
Specify an input ENVIAgCrops object. You can run AgCropCount Task or AgCountAndRasterizeCrops Task to create an ENVIAgCrops object before running this task. Or, you can use an ENVIAgCrops file that you created and saved earlier.
INPUT_RASTER (required)
Specify a multispectral raster that will be used to create the specified spectral index raster.
OUTPUT_CROPS_URI (optional)
Specify a string with the fully qualified filename and path for the output JSON file that will contain the crop data.
OUTPUT_METRIC (optional)
Set this parameter to one of the following strings, indicating the metric that the output raster will be based on:
- Crop Mean (default)
- Crop Maximum
- Crop Minimum
- Crop Standard Deviation
- Radius
OUTPUT_RASTER_URI (optional)
Specify a string with the fully qualified filename and path of the associated OUTPUT_RASTER.
- If you do not specify this parameter, or set it to an exclamation symbol (!), ENVI creates a temporary file.
- If you set it to the hash symbol (#), ENVI creates a file in the temporary directory, but this file will not be deleted when ENVI closes.
Output Parameters
OUTPUT_CROPS
This is a reference to the output crop data.
OUTPUT_RASTER
This is a reference to the output crops classification raster or grayscale raster.
Methods
Execute
Parameter
ParameterNames
Properties
DESCRIPTION
DISPLAY_NAME
NAME
REVISION
TAGS
Version History
|
Crop Science 1.0
|
Introduced |
|
Crop Science 1.1
|
Added Crop Minimum, Crop Maximum, Crop Standard Deviation, and Radius options to OUTPUT_METRIC
|
See Also
AgCalculateCropMetrics Task, AgCalculateAndRasterizeCropMetrics Task, ENVIAgCrops, AgCropCount Task, AgCountAndRasterizeCrops Task